PVA Brain Phantom Images
Simulated
data
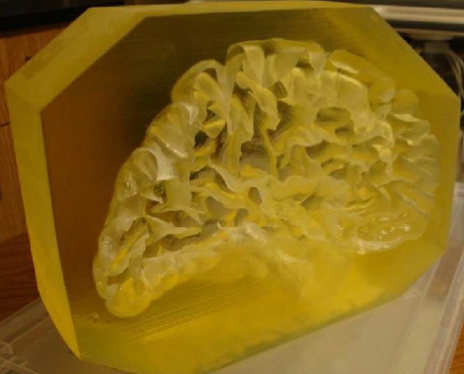
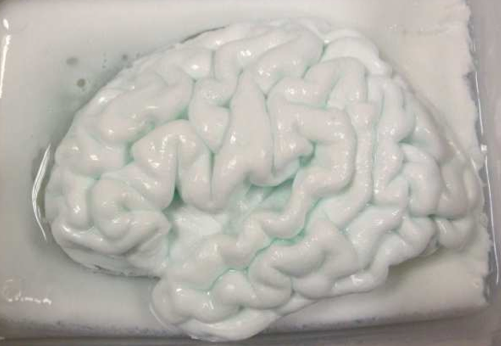
We have created an
anatomically and mechanically realistic brain phantom from polyvinyl
alcohol cryogel (PVA-C) for validation of image processing methods for
segmentation, reconstruction, registration, and denoising. PVA-C is
material widely used in medical imaging phantoms for its mechanical
similarities to soft tissues. The phantom was cast in a mold designed
using the left hemiphere of the Colin27 brain dataset and contains deep
sulci, a complete insular region, and an anatomically accurate left
ventricle. Marker spheres were also implanted to enable good
registration. Inflatable catheters were used to simulate tissue
deformations.
Multi-modality
imaging
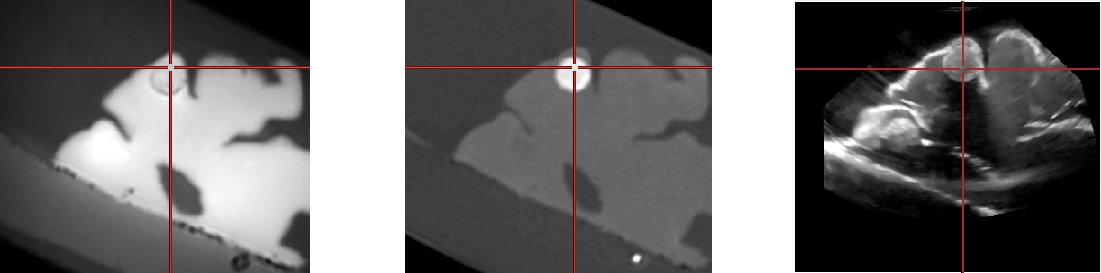
The phantom was designed for
triple modality imaging, giving good contrast images in computed
tomography, ultrasound, and magnetic resonance imaging. Imaging data
from each modality, and from multiple deformation level, were acquired.
This page contains images files of the PVA Brain
phantom. For each deformation instant, you can download one pack that
archives reconstructed and unreconstructed B-mode ultrasound (US)
images at 5.2cm and 7.1cm depth, as well as reformatted MR and CT
volumes. These images are made freely available
to the image processing community.
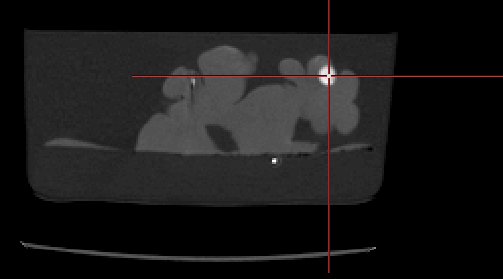
Here is an example
that you can download (about 13 Mo): With no
deformation at all, a first ultrasound volume was acquired (7.1cm
depth, Superior Frontal). MR and CT volumes have been cropped to fit in
this US volume's space.
First line, from left to right: MR T1, CT registered rigidly to MR
Second line, from left to right: Cropped MR, cropped CT and US volume.
The cropped MR and CT were resliced according to the transformation of
the neuronavigation system. Therefore, the volumes are roughly aligned
and not perfectly registered.
All volumes are stored in NIFTI (.nii) format. Download this 5
volume example on the download page (13M).
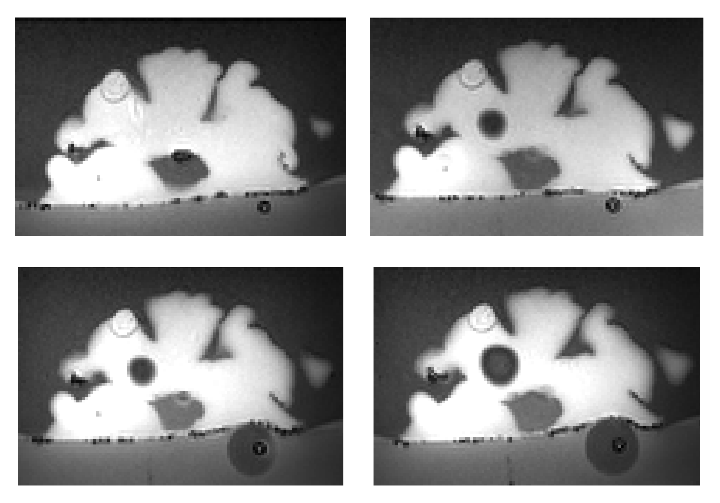
Simulated deformations
Deformations were created as injected
liquid volumes increase in the two catheters. For each deformation
instant, the phantom was imaged with CT, MR and ultrasound.
For each
instant, a image archive containing all images can be
downloaded. Here is
the content of each archive:
- "MR-CT" contains the MR and CT images. The MR sequence
contains a 3D
T1, Flair, T2 and Diffusion weighted sequence.
- "US" contains the acquired ultrasound slices, with different
field of
view. For each sequence, the raw data in stradx format
(http://mi.eng.cam.ac.uk/~rwp/stradx/stradx_files.html) are provided,
as well as the reconstructed volumes in nifty format using the distance
weighted interpolation. For each acquisition, a transformation matrix
(.mat) is also provided that matches the US coordinate system to those
of the "preoperative" MR image.
- "US_denoised" contains the denoised version of the reconstructed US
volumes using the NL-means method.
- "MR_cropped" and "CT_cropped" contain the resliced MR and CT volume
for each US
acquisition. The cropped MR and CT were resliced according to the
transformation of the neuronavigation system. Therefore, the volumes
are roughly aligned and not perfectly registered. The transformation
stored in file US.mat describes the transformation matrix that was used
to reslice the MR according to the FOV of the US. The transformation
was used as an input of the VTKReslice class.
Displacement images
The following are the
images scanned with displacements. Ten of the
scans are done with the phantom submerged in water while the other 10
are done without.
All the displacement images can also be download as
single archive here
(740M).
Reference
and contacts
If you use these images please cite :
Sean
Jy-Shyang Chen, Pierre Hellier, Jean-Yves Gauvrit, Maud Marchal, Xavier
Morandi, and D. Louis Collins (2010) An Anthropomorphic Polyvinyl
Alcohol Triple-Modality Brain Phantom based on Colin27, MICCAI 2010
Contacts
Legal mentions